GrainGenes
A Database for Triticeae and Avena

D. Perovic1, W.D. Smilde1, J. Haluskova4, R. Waugh˛, T. Sasakił and A. Graner1
1) Institute of Plant Genetics and Crop Plant Research (IPK),
˛) Dept. of Cell and Molecular Genetics, Scottish Crop Research Institute (SCRI),
3) Rice Genome Research Program, 446-1 Ippaizuka, Kamiyokoba, Tsukuba,
4) Institute for Resistance Genetics, Graf-Seinsheim-Str. 23, 85461 Grünbach, Germany
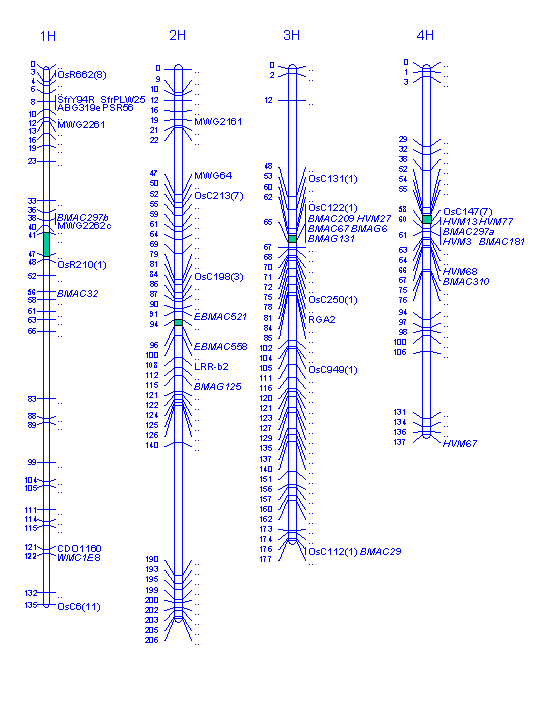
The Igri/Franka map is based on a population consisting of 71 anther-derived doubled-haploid barley plants (Graner et al. 1991, 1993, 1996) and is, except for a major gap on chromosome 2HL, which seems to be due to identity by descent, fairly well saturated with markers. Here, we report on further marker saturation of the Igri/Franka map in the past 4 years. In total, 76 new markers which define 55 map locations (19 new map locations) were added to the map, which comprised previously 477 markers at 211 loci extending over 1214 cM. Most of the new markers are microsatellites. In addition, rice markers as well as resistance gene analogues and some known function genes were mapped. For the sake of clarity, only the newly mapped markers are shown on the map displayed in Fig. 1, while the loci are given without names for previously mapped markers. References for each new marker are given in Table 1. The corresponding raw data and a complete map drawing will be available from the graingenes database (http://wheat.pw.usda.gov).
Microsatellite markers
A total of 151 barley SSR markers were screened for polymorphism between Igri and Franka, on 10 % polyacrylamide gels. Ninety-five of these markers were developed at the SCRI, and most have been integrated in the wide cross Lina x Canada Park. Of these, 30 SSRs (31.6 %) showed polymorphism between Igri and Franka cross and were placed on the map. Primer sequences of these markers are available on request from SCRI. Of another set comprising 45 previously mapped SSR markers (Liu et al. 1996) 14 (31 %) proved to be polymorphic and were mapped in Igri x Franka. Also, four out of 15 SSR markers (26.6 %) described by Becker and Heun (1995), could be placed on the Igri/Franka map. (The numbers don't add up because of some overlap.)
The ability of SSR markers to reveal polymorphism between Igri and Franka hardly exceeded the values observed with a random set of barley RFLP markers. Moreover, SSR markers are not distributed at random across the chromosomes but tend to cluster around the centromeres. Only seven of the 45 mapped SSR markers (16%) were mapped outside the 20 cM regions around the centromeres, while 354/477 (74%) of the previously mapped RFLP markers are out there, so the clustering of SSR markers is stronger than the clustering of RFLPs.
Rice probes
A total of 87 rice probes from the RGP Rice RFLP landmarker set (Antonio et al., 1993) was screened for cross-hybridization with and polymorphism in barley. Twenty-one genomic PstI probes DNA hardly showed any specific cross-hybridization with barley DNA. On the other hand, > 70% of the 60 cDNAs tested revealed specific cross hybridization and 17 (29%) of these were polymorphic and could be mapped. Ten (59 %) of these were mapped a syntenic position as described by Saghai-Maroof et al. (1996). The good overall synteny between rice chromosome 1 and barley chromosome 3H was studied in more detail. The results of this study will be reported separately.
Other probes
In addition to the above-mentioned markers, a series of additional RFLP probes from several sources were mapped, including known function probes, resistance gene analogues of the leucine-rich repeat (LRR) type and some genomic PstI-probes.
The updated map now contains a large number of markers that provide a resource for linkage analysis and for the development of STS markers based on available DNA sequences (Mano et al., 1999; Michalek et al., 1999). Moreover, the integration of a set of anchor markers from both barley and rice facilitates map comparisons within the Triticeae and, although still at a low resolution, between barley and rice.
Table 1. List of 76 new markers placed on the Igri/Franka map (Fig. 1)
| 1H | 2H | 3H | 4H | 5H | 6H | 7H | |||||||
| ABG319e | 6 | MWG2161 | 6 | OsC112 | 4 | OsC147 | 4 | MWG2092 | 6 | MWG286 | 6 | OsC236 | 4 |
| MWG2261 | 6 | MWG64 | 6 | OsC121 | 4 | BMAC181 | 1 | BMAC303 | 1 | cMWG2318 | 6 | OsC356 | 4 |
| MWG2262c | 6 | OsC198 | 4 | OsC131 | 4 | BMAC297a | 1 | BMAC96 | 1 | OsC74 | 4 | BMAC167 | 1 |
| CDO1160 | 6 | OsC213 | 4 | OsC250 | 4 | BMAC310 | 1 | HVDHN9 | 2,3 | OsC174 | 4 | BMAG11 | 1 |
| OsC6 | 4 | BMAG125 | 1 | OsC949 | 4 | HVM3 | 2 | HVLEU | 2,3 | OsC597 | 4 | BMAG110 | 1 |
| OsR210 | 4 | EBMAC521 | 1 | BMAC209 | 1 | HVM13 | 2 | PSR567b | 6 | OsC601 | 4 | BMAG120 | 1 |
| OsR662 | 4 | EBMAC588 | 1 | BMAC29 | 1 | HVM67 | 2 | WG1026 | 6 | BMAC18 | 1 | BMAG21 | 1 |
| BMAC297b | 1 | LRRb2 | 5 | BMAC67 | 1 | HVM68 | 2 | Jip66b | 8 | BMAC297c | 1 | BMAG359 | 1 |
| BMAC32 | 1 | BMAG131 | 1 | HV77 | 2 | Lox1a | 7 | BMAG210 | 1 | BMAG7 | 1 | ||
| WMC1E8 | 1 | BMAG6 | 1 | LRRb8 | 5 | BMAG3 | 1 | HVCMA | 2,3 | ||||
| PSR56 | 6 | HVM27 | 2 | BMAG344 | 1 | HVWAXY | 3 | ||||||
| SfrPLW25 | 9 | RGA2 | 10 | BMAG9 | 1 | HVM4 | 2 | ||||||
| SfrY94R | 9 | BMG1 | 1 | RpoT | 11 | ||||||||
| HVM14 | 2 | ||||||||||||
| HVM22 | 2 | ||||||||||||
| HVM34 | 2 | ||||||||||||
| HVM74 | 2 | ||||||||||||
| Lox1b | 7 | ||||||||||||
| LRR-r11 | 5 |
1 SSR from the Scottish Crop Research Institute, Dundee, United Kingdom
2 SSR described by Liu et al., 1996
3 SSR described by Becker and Heun, 1995
4 Rice cDNA marker described by Harushima et al., 1997
5 LRRs described by Leister et al, 1998
6 Locus/probe source described in Graingenes (http://wheat.pw.usda.gov)
7 Lox=lipoxygenase, Vörös et al., 1998
8 Jip=Jasmonate induced protein, C. Wasternack, Institut für Pflanzenbiochemie, Halle Germany
9 Feuillet and Keller, 1999. Sfr=Swiss Federal Research
10LRR-RGA, T. Börner, Humboldt-Universität, Berlin
11 RNA polymerase, W.R. Hess, Humboldt Universität, Berlin

References
Antonio, BA, Yamamoto K, Harushima Y, Sue N, Shomura A, Lin SY, Inoue T, Fukuda A, Shimano T, Hayasaka H, Kuboki Y, Zungsontion S, Yano M, Kurata N, Nagamura Y, 1999. More than one thousand markers mapped by RFLP. Rice Genome 2 (2): 3-8
Becker J, Heun M, 1995. Barley microsatellites: allele variation and mapping. Plant Mol Biol 27: 835-845
Feuillet C, Keller B, 1999. High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci U S A, 96 (14): 8265-70
Graner A, Jahoor A, Schondelmaier J, Siedler H, Pillen K, Fischbeck G, Wenzel G, Herrmann RG, 1991. Construction of an RFLP map in barley. Theor Appl Genet 83: 250-256
Graner A, Bauer E, Kellermann A, Kirchner S, Muraya JK, Jahoor A, Wenzel G, 1993. Progress of RFLP-map construction in winter barley. Barley Genetics Newsletter 23: 53-59
Graner A, 1996. RFLP mapping the haploid genome of barley (Hordeum vulgare L.) In: Jain SM, Sopory SK, Veilleux RE (eds.) In Vitro Haploid Production in Higher Plants, Kluwer, Dordrecht/Boston/London Vol. 2: 127-150
Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, Kajiya H, Huang N, Yamamoto K, Nagamura Y, Kurata N, Khush GS, Sasaki T, 1997. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148: 479-494
Künzel G, Korzun L, Meister A, 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154: 397-412
Leister D, Kurth J, Laurie D, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P, 1998. Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA 95: 370-375
Liu ZW, Biyashev MR, Saghai Maroof MA, 1996. Development of simple sequence repeat DNA markers and their integration into a barley genetic linkage map. Theor Appl Genet 93: 869-876
Mano Y, Sayed-Tabatabaei BE, Graner A, Blake T, Takaiwa F, Oka S, Komatsuda, T, 1999. STS marker-based map construction in Japanese barley (Hordeum vulgare L.). Theor Appl Genet 98: 937-946.
Michalek W, Künzel G, Graner A, 1999: Sequence analysis and gene identification in a set of mapped RFLP markers in barley (Hordeum vulgare). Genome 42: 849-853
Saghai-Maroof MA, Yang GP, Biyashev RM, Maugham PJ, Zhang Q, 1996. Analysis of the barley and rice genomes by comparative RFLP linkage mapping. Theor Appl Genet 92: 541-551
Vörös K, Feussner I, Kühn H, Lee J, Graner A, Löbler M, Parthier B, Wasternack C, 1998. Characterization of a methyljasmonate-inducible lipoxygenase 2 from barley (Hordeum vulgare cv. Salomé) leaves. European Journal of Biochemistry 251: 36-44
